Heather Louise Helton, MD
 Heather Louise Helton, MD
Heather Louise Helton, MD
Fred Hutchinson Cancer Research Center
Biologic and Clinical Implications of ETV6 Genomic Variations in AML
My research aims to improve treatment for children with acute myeloid leukemia (AML), a type of blood cancer. My work is focused on characterizing the scope of mutations in the ETV6 gene in children with AML. My preliminary work indicates that patients with ETV6 mutations are more likely to have a poor outcome. I hope to use the mutations in ETV6 as a marker to identify patients at high risk of relapse at the beginning of their treatment in order to predetermine therapies that are most likely to succeed.
Grant Term: July 1, 2013 – December 30, 2014
Research Summary:
My work is aimed to move towards personalized medicine that targets cancer therapy to each individuals disease. I have worked to characterize genetic changes in ETV6 and what results these changes have in acute myeloid leukemia in children. Eventually, my hope is to identify novel drug therapy for patients with leukemia whose cancer cells contain these genetic alterations to help improve outcomes and limit toxicity.
Research Progress
Specific Aims
1. Determine the full spectrum of ETV6 genomic alterations (mutations, deletions, translocations) in a large cohort of pediatric and adult AML.
2. Define the association of ETV6 variants with specific disease characteristics and clinical outcome.
3. Determine the biologic implications of ETV6 alterations by devising an in-vitro system to study the cellular implications of specific ETV6 alterations as well as evaluate the associate mRNA and miRNA expression
profile of ETV6 variants in primary patient samples.
Results obtained to date
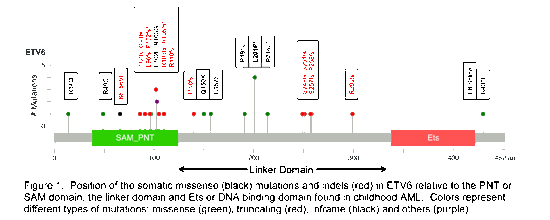
Mutations: In regards to determining the full spectrum of ETV6 genomic alterations, targeted sequencing of ETV6 is now complete for 1092 diagnostic specimen from 2 consecutive de novo childhood AML treatment protocols, COG AAML03P1 and 0531. Fragment length analysis for an additional 150 patients from AAML0531 is also complete. In total, 30 somatic mutations were identified in 26 patients including 14 missense, 1 splice site mutation and 15 indels (14 frameshift and 1 in-frame) as documented in Figure 1. Of the missense mutations, 71% are predicted to be probably or possibly damaging by Polyphen-2 and 73% of indels result in a premature stop codon per in silico translation. One patient had two missense mutations and another patient had two consecutive point mutations and two 2-bp deletions confirmed by repeat sequencing and found on a single allele after cloning of the product.
A majority of the identified mutations, including truncating mutations, are clustered around the HLH domain of ETV6 important for formation of the HLH structure, homo-dimerization and interactions with other co-factors. Another hotspot is in the linker domain that is also important for dimerization and influences DNA binding. Mutations in these areas may lead to an altered protein product and ETV6 transcriptional repressor activity.
Deletions: Nanostring CNV assay in 212 diagnostic samples from patients with childhood AML identified 2 patients with intragenic deletions: one patient with Exon 5 deletion and one patient with deletion of Exons 6-8. An additional 2 patients had deletion of the entire gene and a large portion of the flanking upstream and downstream region and available cytogenetic data revealed translocations involving 12p13 involving ETV6.
Translocations: As described in my grant proposal, translocations involving ETV6 and Nom-1, INO80D and PDGFRB were identified by WGS from TARGET AML Initiative. I screened 160 patients for INO80D/ETV6 and HLXB9/ETV6 fusion. HLXB9 has been identified as the partner in t(7;12) in contrary to previous reports implicating Nom-1. Two samples with known t(7;12) were positive for HLXB9/ETV6 but none of the samples tested were positive for INO80D/ETV6 fusion leading me to conclude that translocation events involving ETV6 are rare in childhood AML.
Statistical analysis of the above results is currently ongoing in collaboration with COG statistician, Todd Alonzo per Aim 2 of the project.
ETS family: In addition to ETV6, I have started to expand my research to investigate genomic alterations in other ETS family transcription factors based on TARGET data. I verified point mutations in ERF and SPI1 by direct sequencing and fusions involving ERG and FLI by RT-PCR. Deletion of ELF1 was identified in 15% (N=32) out of 212 patients by Nanostring CNV assay.
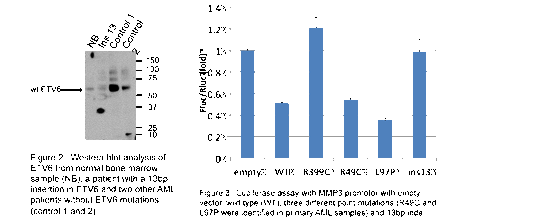
Functional studies: As noted above, in silico translation predicts a truncated ETV6 protein product in a majority of the identified alterations. I confirmed a truncated protein product in a patient with a 13 bp deletion involving exon 3 of ETV6 as predicted [Figure 2]. Curiously there was very little, to no wt ETV6 in this patient, perhaps suggesting a possible mechanism whereby mutated ETV6 suppresses expression of wt ETV6.
In collaboration with the Shimamura lab at FHCRC, I introduced point mutations and indels to model the types of alterations found in primary patient samples using a mutagenesis kit and transiently transduced HELA cells with pHAGE ETV6 mutants. Luciferase assay testing of empty vector, wild type, positive control (R399C), two ETV6 point mutations (R49C and L97P) and one indel (ins13) found in primary AML patient samples revealed similar transcriptional repression activity for the point mutations compared to WT and loss of function for the indel [Figure 3]. My next steps will be to complete localization studies to determine if any of the mutants result in impaired nuclear localization.
Upon completion of these additional functional studies and statistical analysis, I plan to publish my results.
Mission Statement
The Strike 3 Foundation heightens awareness, mobilizes support, and raises funding for childhood cancer research.
How you can help
Contact Us
Strike 3 Foundation
PO Box 191
Monroe, CT 06468
(203) 724-1067